The following is an experiment of features in Tableau, and is not a presentation of normalized compete data for the Flora of Michigan.
The following is a test of using the
Tableau program to explore a collection of
Herbarium resources at the
University of Michigan. These data have not been normalized and do contain some inaccuracies. The species indicated as invasive are based on a query that does contain some problems.
For an accurate and verified Flora of Michigan please see the Flora of Michigan web site which includes an accurate vetted assessment of alien status in Michigan. The new Field Manual of Michigan Flora is also a must-have for anyone interested in the Flora of Michigan.
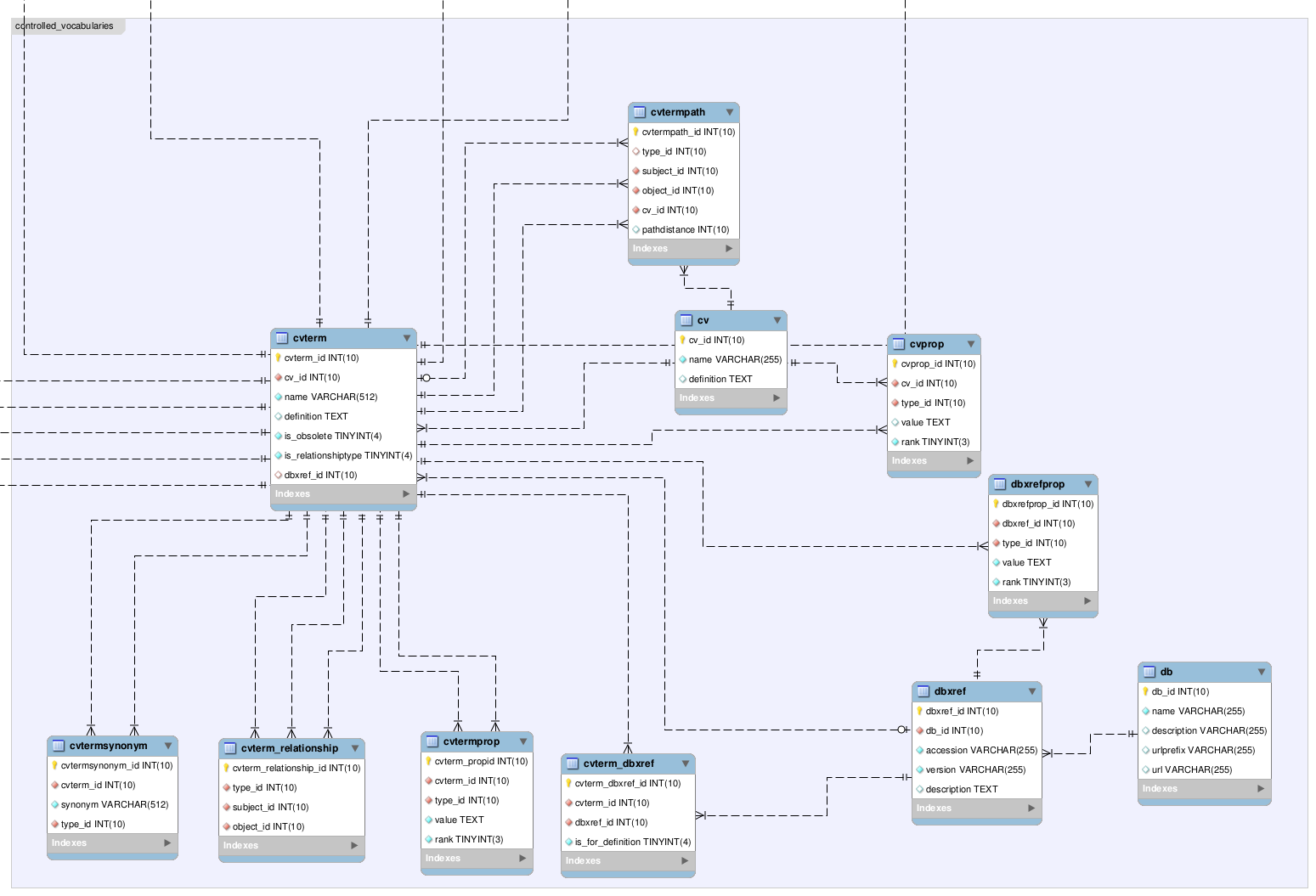
The Tableau software allows us to connect to a live database such as
MySQL or
MS Access, and create informative visualizations. These visualizations can be exported as images, pdfs, and even presented on the web as interactive worksheets. In the example below, I connected to a summary table in an MS Access database that was converted to a
Tableau data extract for optimized queries. These data extracts can be updated from the live data on a regular basis either manually or in an automated fashion. This allows for fully optimized read-only access to the data, while allowing for regular updates to reflect the current state of knowledge for a collection.
Overview of a Michigan Herbarium Database
Using Tableau it is possible to capture a tremendous about of information about a museum collection in a single view. For example, taking a database of herbarium specimens as input, the image below is an overview of the collections of native and invasive species collected in Michigan. This shows the county level distribution of the collection and provides an the indication of when the taxon was first collected in the state.
This view summarizes nearly a quarter of a million specimens, and has multiple parts summarizing this data.
Map and Summary
The upper right hand area of the view shows a map of the distribution of accessions and a summary for that view.
The upper right hand panel shows a summary of the number of distinct taxa names, the number of distinct collector names, and the total number of accessions in the collection. The current view shows 8,432 distinct taxon names in the collection occur in the state of Michigan. This number includes some misspellings as well as many subspecies, varieties and named hybrid species. There are over 6,000 collector names that have accession in the collection, but this is currently an overestimate since it is doing a match on the full name of the collector. Misspellings and accessions with multiple collectors are inflating this number.
The size of the dots over each county represent the number of accessions for that county, with putative invasive species shown in orange and all other species indicated in blue.
Collection Years
The bottom panels of the view explores the temporal pattern of the collection.
The bar chart at the top represents the number of state records that are recorded for that year. These are the first known
vouchered occurrences for those species in the state of Michigan. Other spottings may have been recorded, but these records are physically represented in the collection. Similar to the distribution map above, this chart shows putative invasive species in orange and all other species in blue.
The bar chart at the bottom represents the total number of accessions collected in that year. Comparing the top chart to the bottom illustrates that many of the important early state records were some of the only specimens collected in that year. We can also clearly see the
effect of World War II on collecting in Michigan.
List of Taxa
The top middle panel gives the list of distinct taxon names.
This is a scrollable list that gives the number of accessions for that taxon in the state. Putative invasive species are shown in orange and all other species are indicated in blue.
"Acer ruburm an invasive!" you say ..
yes this is native in Michigan. See ... I told you this is an illustration of a process that can be facilitated by Tableau, not publishable hard science. One of the things that Tableau facilitates is finding errors and outliers in your data set that can be fixed by data normalization before taking your findings to print.
Who did all the Work
The top left panel is a bubble plot of the scientists that have contributed to the collection.
The larger the dot the more vouchered specimens the individual biologist have contributed to the collection. In the interactive view, hovering over an individual dot reveals the name of the scientists and the number of accessions in the collection.
Filter Records by Subcollection
The real power in the Tableau interface is the interaction among the panels. For example, we can select what components of the collection we want an overview for by selecting the sub-collections in the panel on the right.
Bryophytes (Mosses)
Vascular Plants
It is possible to show any combinations of the subcollections. For example the Monocots, Dicots, Gymnosperms and Pteridophytes.
Fungi
The following shows only the fungi records for Michigan in the database.
Filter Record by Location
All collections from Washtenaw County
It is also possible to use a view in one panel to filter the data displayed on other panels. For example, selecting Washtenaw county the location of the University of Michigan will show the collection statistics for that county. The plot of the year the taxon was first collected now represents vouchered county records for Washtenaw county. The taxa list is the list of all species known to have been collected in Washtenaw county.
Washtenaw County Native Species
Selecting on the blue portion of the pie in Washtenaw county will only display data for putative native species in Washtenaw county.
Washtenaw County Putative Invasive Species
Similarly selecting only the orange part of the pie will show data for putative invasive species in Washtenaw county. Again the year of first occurrence is the first year that putative invasive species was collected from Washtenaw county.
All Records from the Michigan Upper Peninsula
We can spatially select multiple counties as well. For example, we can limit the data display to all records in the Upper Peninsula of Michigan. The year first collected bar chart will show the year the species was first collected in the UP, and the taxa list is the list of all species collected in the UP.
Filter Records by Taxonomy
Fagaceae
I like the Oak family so we can limit a display to the Fagaceae.
Quercus
Limiting to all species of Quercus.
Quercus bicolor
We can even show just a single species, Quercus bicolor while ignoring the named hybrids.
Contributions of Single Individuals
Ed Voss
For example we can show the collections from
Ed Voss in Michigan. The county map represents his individual contributions to the flora of Michigan, and the Year First Collected panel represents the first time he personally collected that species in the state of Michigan.
Fungus Collections from A.H. Smith
We can combine a filter on sub-collection and collector to visualize the fungus collections from
Alexander H. Smith. This illustrates the diversity of his collecting, and shows that much of his work was conducted in the vicinity of the
University of Michigan campus in Ann Arbor, and the
University of Michigan Biological Field Station at the northern tip of the Upper Peninsula.
Why I like Tableau
As you can see, Tableau provides an exciting way to view natural history collections that adds a level of interactivity to a traditional distribution dot map. These views can help highlight needs in existing collections, and can provide an interactive way to highlight any accuracy issues in the underlying data.
Tableau provides a view of the biodiversity of a region that is anchored in vouchered specimens, and will hopefully serve as a venue for gaining new biological insights from a collection. Once data are available in an interactive form on the web, users will have access to on-demand lists and reports that will add value to any museum collection database.